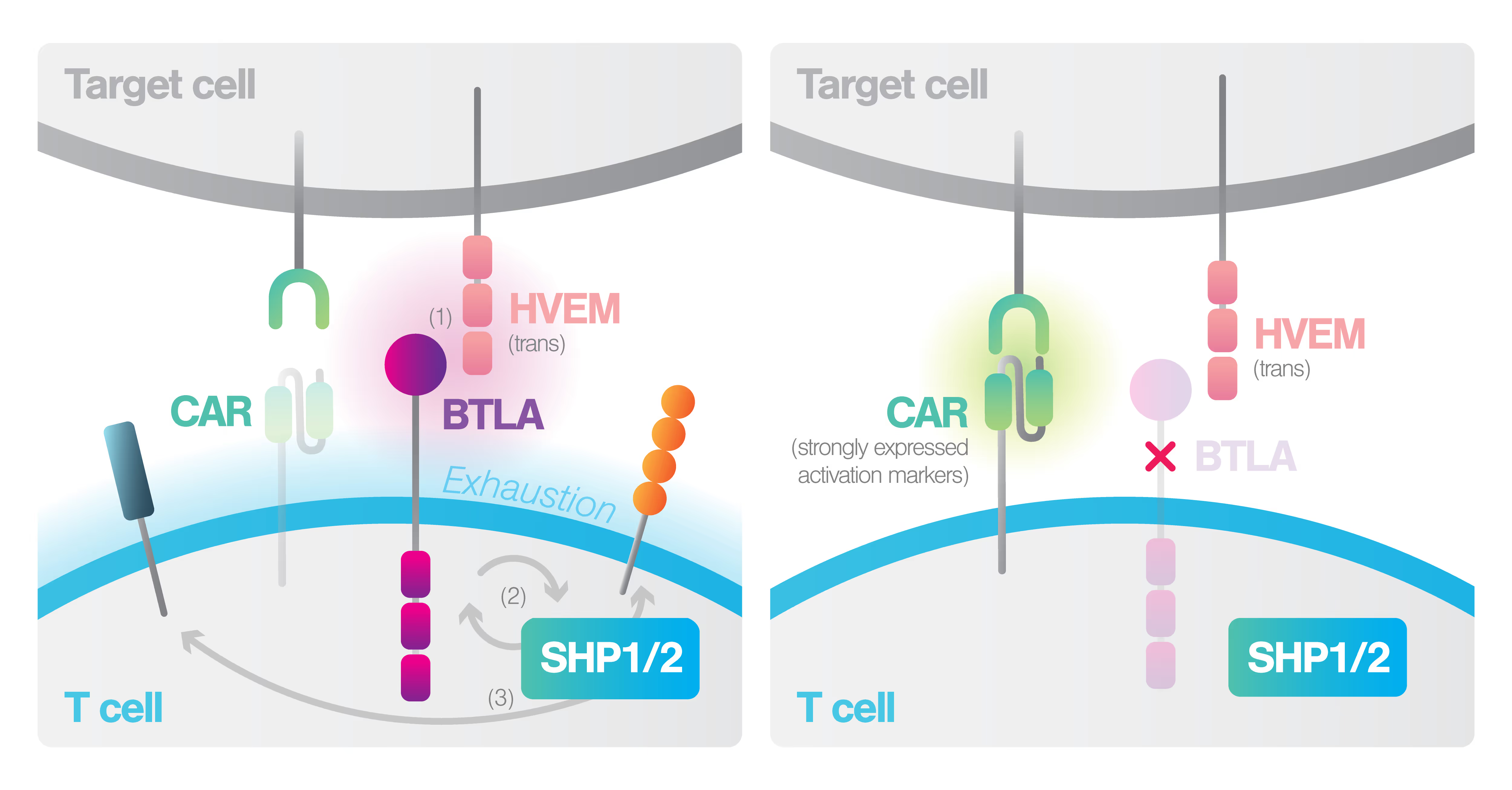
Improving CAR-T immunotherapy for lymphoma by fine-tuning avidity
Abstract
Commercial CAR-T therapies still suffer from severe limitations, as majority of patients fail to achieve complete response and ultimately relapse.
Watch this Webinar to discover Prof. Marco Ruella’s team have adopted a novel CAR-T avidity screening method to improve safety and exhaustion profile, leading to 100% clinical response in a phase I trial.
What you'll learn
- How an innovative CAR binder leads to enhanced clinical response compared to currently available commercial CAR.
- How tuning the fine balance between T cell engagement and exhaustion in CAR T cells leads to a more durable anti-tumor response and superior clinical outcome.
- The innovative strategy used to reduce Activation-Induced Cell Death (AICD) in CAR T cells, a key challenge in providing durable and complete response for patients.
Get 5 of each and filter based on business unit client side.
This shows 5 of each card, and then filters them client side on BU.
Webinar, Scientific update, Whitepaper, Application note, Brochure.
We only show 4 cards in the end.
In design only 1 is shown, but the rest will be loaded when published.
Tuning CAR-T cells by targeting cancer-associated glycan in pancreatic cancer
Chimeric antigen receptor (CAR) T cell therapy has transformed cancer treatment, but its efficacy remains limited in solid tumors due to antigen heterogeneity, an immunosuppressive microenvironment, and the glycocalyx barrier. The glycocalyx, composed of dense glycoproteins such as MUC1, is markedly expanded in cancers, where it impedes immune cell access and antigen engagement, thereby reducing therapeutic efficacy. In most adenocarcinomas, the Tn antigen, comprising N-acetylgalactosamine linked to serine or threonine, is overexpressed. Tn-MUC1, a truncated form of MUC1 decorated with Tn antigen, is frequently overexpressed in pancreatic cancer. Here, we incorporate a non-signaling glyco-bridge binder recognizing Tn-MUC1 into mesothelin-directed CAR-T cells. This bridge enhances tumor recognition and cytotoxicity by increasing avidity and facilitating CAR activation in a density- and affinity-dependent manner. To directly validate these effects at the cell interaction level, we used Lumicks z-Movi to quantify CAR-T binding strength to tumor targets. CAR-T cells equipped with the Tn-MUC1 glyco-bridge exhibited higher cell avidity toward Tn-MUC1-expressing tumor cells compared to a CD19 bridge control. To broaden its applicability, we design a tandem Helix pomatia agglutinin (HPA) lectin-based bridge that recognizes Tn antigens across cancer types. CAR-T cells with the HPA-bridge exhibit superior cytotoxicity in pancreatic cancer models.
Improving the anti-tumor efficacy of CAR-T through biomolecular condensation
Chimeric antigen receptor (CAR)-T cell therapies showed remarkable efficacies in treating otherwise intractable cancers. However, current clinically approved CAR-T therapies are limited by low antigen sensitivity, impeding their efficacy against cancers with low antigen expression. Here, to address this issue, we engineered CARs targeting CD19, CD22 and HER2 by including intrinsically disordered regions (IDRs) that promote signaling condensation. We discovered that the CAR fused with an IDR from FUS, EWS or TAF15 promoted the formation of CAR-T conjugation with cancer targets, the mechanical strength of CAR-T synapses and membrane-proximal signaling, which led to an increased release of cytotoxic factors and a higher killing activity toward low-antigen-expressing cancer cells in vitro. Moreover, the IDR CAR-T displayed improved antitumor effects in both blood cancer and solid tumor models. No elevated tonic signaling was observed. Together, our work demonstrates IDRs as a new toolset for improving CAR-T function through inducing biomolecular condensation.
This webinar is in Chinese. 此次线上研讨会为中文讲座
Cell Avidity Uncovers Novel KIR:HLA binding predicting patient survival post stem cell transplant
Despite significant progress in haploidentical hematopoietic cell transplantation (haploHCT) for leukemia, relapse-free survival remains a major challenge, even with the addition of immune effectors such as NK or T cells. Given the crucial role of NK cells and their killer immunoglobulin receptors (KIRs) in early anti-leukemic activity, this study sought to refine the understanding of KIR:HLA (human leukocyte antigen) interactions in order to better predict patient outcomes.Using a combination of in silico protein folding and interaction modeling with in vitro acoustic force microscopy to quantify immune synapse avidity, Peter's group identified and validated a novel functional interaction between full-length KIR2DS4 and HLA-B*35. This interaction was confirmed through cell avidity assays with monoallelic KIR and HLA cell lines. When applied to clinical data, patients who received haploHCT and NK cell addback from donors expressing only full-length KIR2DS4 showed significantly improved overall and relapse-free survival. These findings were independently validated in an adult cohort, revealing consistent survival advantages.This newly defined KIR2DS4:HLA-B*35 axis represents an immediately actionable marker for donor selection and offers a mechanistic insight into optimizing NK cell-mediated anti-leukemic responses.
Rational CAR design: Integrating affinity and tonic signaling
The success of chimeric antigen receptor (CAR) T cell therapy for hematological malignancies has not yet translated into long-term elimination of solid tumors, indicating the need for adequate tuning of CAR T cell functionality. The CAR binding moiety is the critical trigger for CAR T cell signaling. CAR binding affinity alone does not determine T cell effector functions. In a panel of anti-Her2 CARs covering a 4-log affinity range, we observed that rather high affinity and cell avidity above the minimum threshold, combined with elevated tonic signaling, produce adequate T cell capacity for expansion and tumor control. The same scFv mutations increased both antigen-specific affinity, cell avidity, and antigen-independent tonic signaling; above a minimum threshold, raise in affinity translated into cell avidity in a non-linear fashion. In this case, replacement by amino acids of higher hydrophobicity within the scFv coincidentally augmented affinity, non-specific binding, spontaneous CAR clustering, and tonic signaling, all together relating to T cell functionality in an integrated fashion. Data highlight the mechanistic complexity of CAR signaling and suggest inclusion of additional variables, for example, hydrophobic interactions, into the equation when determining the CAR’s antigen-specific and tonic signaling capacities.
Enhancing efficacy against clear cell renal cell carcinoma through format-tuning of bispecific T cell engagers
Cell Avidity uncovers critical binding mechanism impairing T-cell potency in the Tumor Microenvironment
Cell Avidity screening deployed by T cell pioneers to accelerate the identification of functional TCRs
Cell Avidity-tuning ensures safe CAR-T therapy for solid tumors
Cell Avidity: a key to accelerate IND filing in cell therapy drug development
Accelerate your cell engager discovery with high throughput measuremenets of Cell Avidity
Accelerate your cell engager discovery with high throughput measurements of Cell Avidity
T cells play a pivotal role in tumor immunosurveillance. Multispecific cell engagers (CEs) have been adopted in the field of immuno-oncology to redirect T cells toward cancer cells, thereby unleashing the anti-tumor potential of the patient’s immune system. CE-mediated cell binding induces T cell activation and the formation of an immunological synapse, which is a prerequisite for effective tumor cell lysis.
The strength of the initial binding events between a T cell and a tumor cell dictates the efficiency of the anti-tumor response. Assessing cell avidity, i.e. the total intercellular interaction strength between two cells, gives crucial insights into the efficacy of CEs as anti-tumor therapeutic agents.
Here, we deploy LUMICKS’ high throughput avidity measurement (HTAM) technology to measure CE-induced cell avidity in a high throughput manner. We demonstrate the assay performance characteristics, i.e. specificity, precision, and range, via CE titration experiments in the context of a Jurkat T cell model system. We find that the HTAM CA assay is suitable for candidate screening in high throughput, with high sensitivity and precision.
Identify functionally optimal TCR T cells through Cell Avidity measurements
T cell receptor (TCR) -based cancer immunotherapy has the potential to become a powerful approach to treat solid tumors, such as melanoma. However, conventional methods that validate the effectiveness of TCR transduced T cells are often inconsistent with functional assays or are tedious to perform. In this application note, we show how the z-Movi® Cell Avidity Analyzer reliably and quickly identifies functionally optimal TCR-engineered T cells targeting melanoma cell lines based on cell–cell interaction strength (cellular avidity).
Rapid assessment of CAR T cell strategies for multiple myeloma with the z-Movi Cell Avidity Analyzer
Identifying effective immunotherapeutic treatment strategies for multiple myeloma that also mitigate relapse often requires tedious and time-consuming validations, such as cell-killing assays and in vivo engraftments. We show that intercellular binding strength (cell avidity), measured by the z-Movi® Cell Avidity Analyzer, quickly predicts CAR T-cell efficacies that correlate with treatment outcomes in vivo.