The mechanical forces driving cellular function and fate
Kinetochore-mediated chromosome segregation
In this experiment, a microtubule was immobilized to a biotinylated coverslip, and Ndc80 complexes, a key protein complex in the kinetochore structure, were attached to streptavidin-coated beads. By moving the bead with the C-Trap, we generated a rupture force between the microtubule and the kinetochore structure.
Researchers from David Barford’s team quantitatively assessed the outer kinetochore’s force resistance of the Dam1 and Ndc80 complexes. By comparing rupture forces of various mutations, they get a better understanding of which protein domains influence the kinetochore’s ability to withstand microtubule forces during cell division.

Dive into the publication
Cellular tension propagation revisited
Measuring membrane tension is challenging as it requires precise manipulation and measurement of mechanical forces on individual tethers, which are small and difficult to control. With the C-Trap this could be resolved by combining optical tweezers for direct measurement of mechanical forces on individual tethers with optogenetics for local endogenous control of cell protrusion.
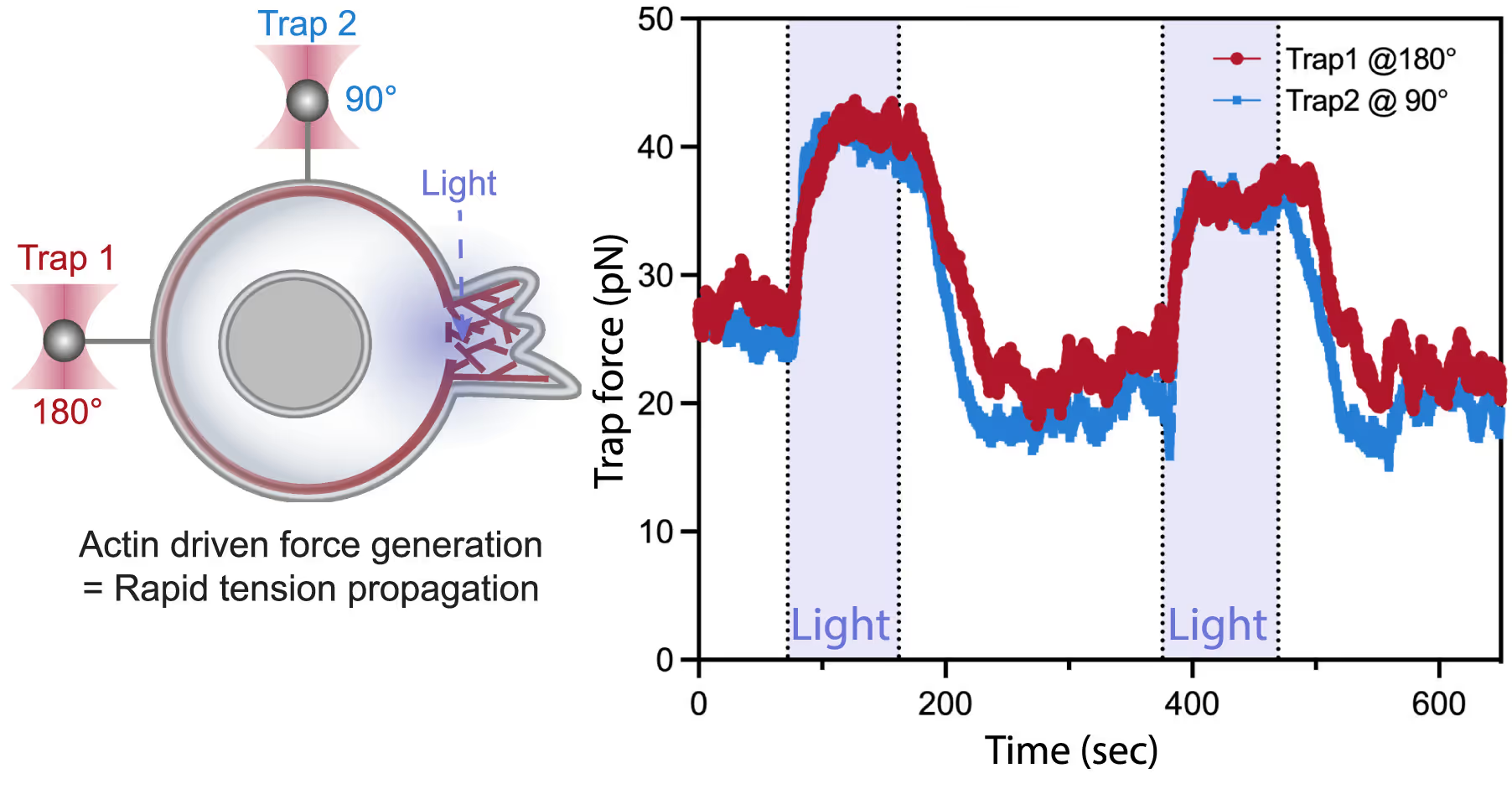
By using this combination of techniques, the lab of Bustamante was able to directly measure changes in membrane tension in response to mechanical forces generated by actin-driven protrusions. This allowed them to demonstrate that these protrusions generate rapid long-range membrane tension propagation in cells, which provides new insights into how cells respond to external stimuli and adapt to their environment.
Left: A dual-tether pulling assay to simultaneously monitor membrane tension on the far end (left, trap 1 at 180°) and on the side of the cell (top, trap 2 at 90°) during light-activated protrusion. Right: Representative time traces of dual trap forces over successive cycles of light-activated protrusion show coinciding tension increases on both membrane tethers adjacent to (trap 2) and at the opposite cell surface from (trap 1) protrusion; light: 90 s on (shaded area), 180 s off. Figure from De Belly et al. (2023), Cell
Dive into the publication

Live cell force dynamics – do cell membranes support or resist tension propagation?
What are the rules of membrane tension propagation in cells? For proper function, cells need to link short-range biochemical signaling events with long-range integration of cell physiology. Forces transmitted through the plasma membrane are thought to serve as this globally integrator. However, conflicting observations have left the field divided as to whether cell membranes support or resist tension propagation. This discrepancy likely originates from the use of exogenous forces that may not accurately mimic endogenous forces. We overcome this complication by leveraging optogenetics to directly control localized actin-based protrusions or actomyosin contractions while simultaneously monitoring the propagation of membrane tension using dual-trap optical tweezers. Surprisingly, actin-driven protrusions and actomyosin contractions both elicit rapid global membrane tension propagation, whereas forces applied to cell membranes alone do not. We present a simple unifying mechanical model in which mechanical forces that engage the actin cortex drive rapid, robust membrane tension propagation through long-range membrane flows.
Cellular droplet fusion and small components in a multicellular organism
Using brightfield microscopy, we can quickly scan cell samples with a micrometer-precision stage and, on top of that, obtain fine movements when needed using a nanometer-precision positioning system. These features allow us to optically trap droplets in two ways: dragging the droplets around the body, thus exposing them to each other, or keeping them at the same position and letting the stage bring other droplets into the proximity of the trapped molecule.
The approach allowed us to follow the small components in vivo and in real time and assess forces associated with the fusion process (Figure 3).
Sample courtesy of Prof. Dr. Daniel Starr at the University of California, Davis
Dive into the publication
C-Trap
Molecular biology as never seen before
The C-Trap® provides the world’s first dynamic single-molecule microscope to allow simultaneous manipulation and visualization of single-molecule interactions in real time.

These cards are NOT components because they use the finsweet nested collection logic. To pull in the posible multiple people wo worked on it.
This type of 1-many relation is not supported native in Webflow.
Also this section is hidden when emtpy. To keep everything visible here, that is being done outside the webflow designer from within Slater.
A Versatile GPMV-Imaging Platform for Quantitative Analysis of Receptor Binding and Membrane Fusion
Migrasome formation is initiated preferentially in tubular junctions by membrane tension
Single Cell Reactomics: Real‐Time Single‐Cell Activation Kinetics of Optically Trapped Macrophages
SITC 2025
CAR-TCR Summit 2025
CICON 2025