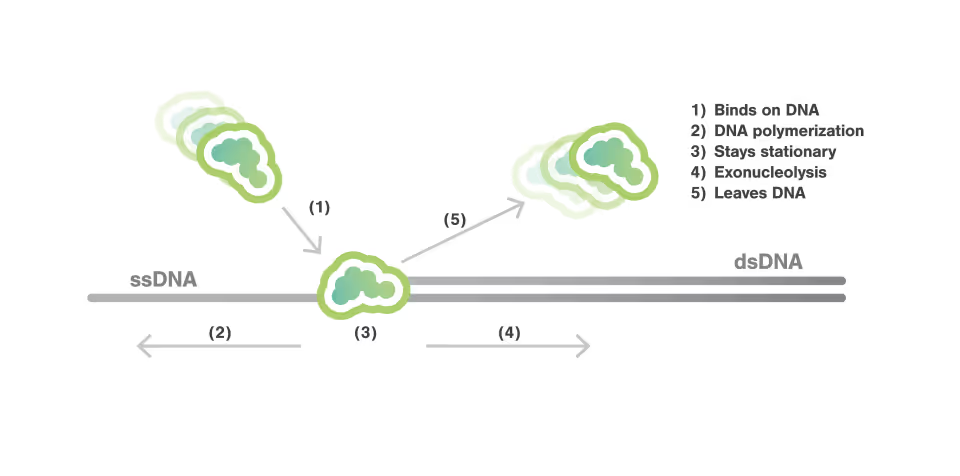
The complex mechanisms inside of the replisome
- Visualize single proteins in real-time
- Assemble the replisome step-by-step
- Manipulate the biological system
Shed light on the role of SMC5/6 in regulating replication fork stability
Eukaryotic DNA replication is one of the most complex molecular mechanisms known. Many of the molecular components serve multiple purposes to maintain the delicate balance between unwinding DNA and synthesizing strands simultaneously while ensuring structural stability and genome integrity.
Dynamic Single-Molecule microscopy, the lab of Shixin Liu (The Rockefeller University New York) was able to simultaneously visualize how Smc5/6 acted on single stranded (ss) and double stranded DNA (dsDNA).
Single molecule fluorescence and force microscopy revealed that:
- Smc5/6 can move over dsDNA in a dynamic manner
- It binds stably to protect junction DNA by preventing ssDNA annealing
Characterization of this previously unknown multifaceted DNA association behavior of Smc5/6 offers a framework for comprehending its various roles in viral DNA restriction and genome maintenance.
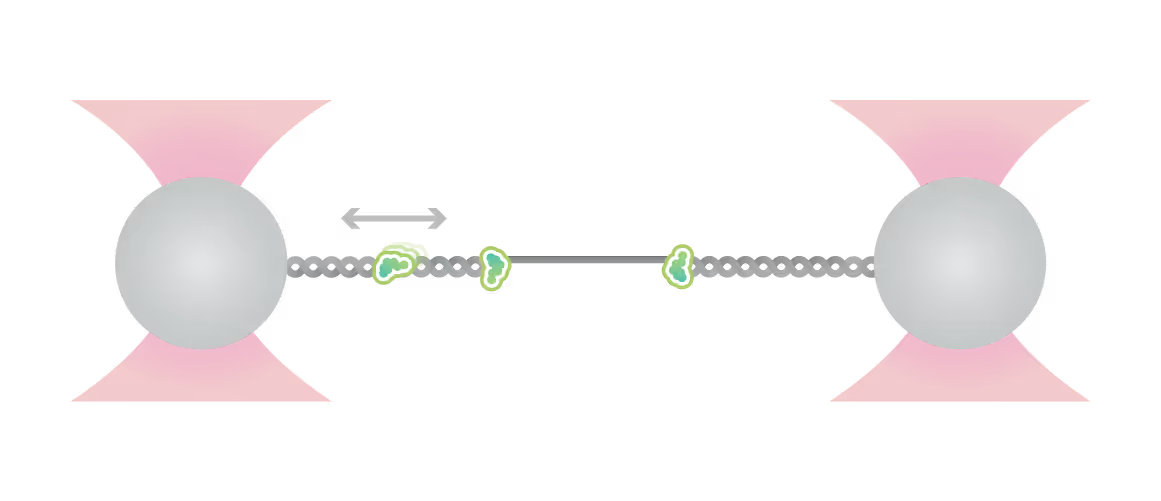
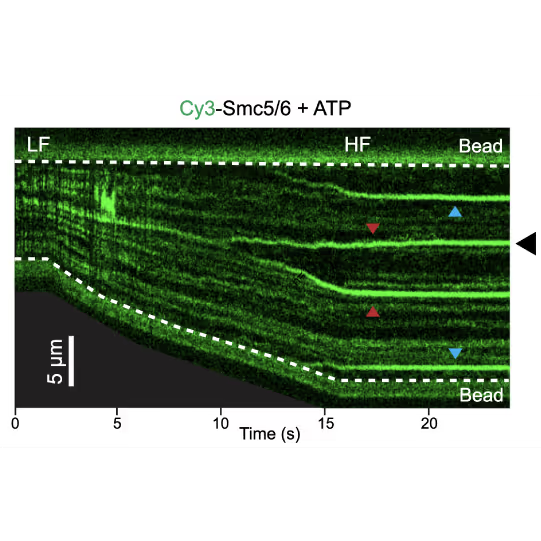
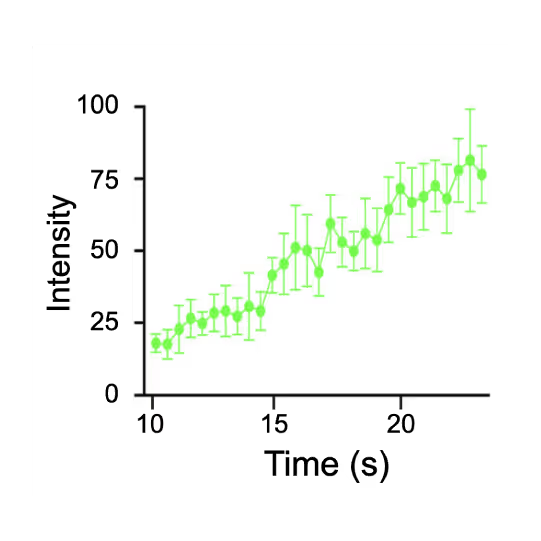
Representative kymograph of a DNA tether in the presence of 20 nM Cy3-Smc5/6 (green) and 2 mM ATP. Force-induced DNA melting creates ssDNA-dsDNA interfaces (forks). Red arrows indicate twin-streaks emerging from an internal DNA nick, and blue arrows indicate single streaks emerging from untethered DNA termini.
Dive into the publication

Breakthroughs into molecular machines on DNA and chromatin
Genome replication and gene expression are carried out by macromolecular machines that exist at nanometer scale and generate piconewton forces. Challenged by the hierarchical chromatin organization and omnipresent thermal fluctuations, these DNA-based machines still accomplish their tasks with remarkable efficiency and accuracy. We leverage single-molecule techniques, particularly correlative fluorescence and force microscopy (smCFFM), to probe the dynamics and mechanics of replication, transcription, and chromatin machinery. These investigations have yielded new insights into the principles of genetic and epigenetic inheritance.
Real-time insights into origin recognition and replisome formation
Understanding the steps and factors that influence originrecognition and licensing is a key challenge in DNA replication research. Modelsystems like yeast tend to exhibit different behavior compared to othereukaryotic cells which makes it difficult to develop a uniform model.
Being able to directly observe loading and dynamic movement of origin recognition complexes and other molecular players in the context of nucleosomes and bare DNA, the labs of Shixin Liu and Nynke Dekker were able to identify and quantify previously unknown fundamental mechanisms via DSM microscopy:
- ORC performs dynamic search on DNA and colocalizes with origin sequences
- ORC recruits MCMs, leading to intermediate complex formation with distinct dynamic behavior
- The presence of nucleosomes has a stronger impact on ORC and MCM binding than sequence
These and other findings enabled by DSM microscopy have fundamentally driven our understanding of replication initiation forward, also closing a gap between different mechanisms of origin recognition and replisome formation.
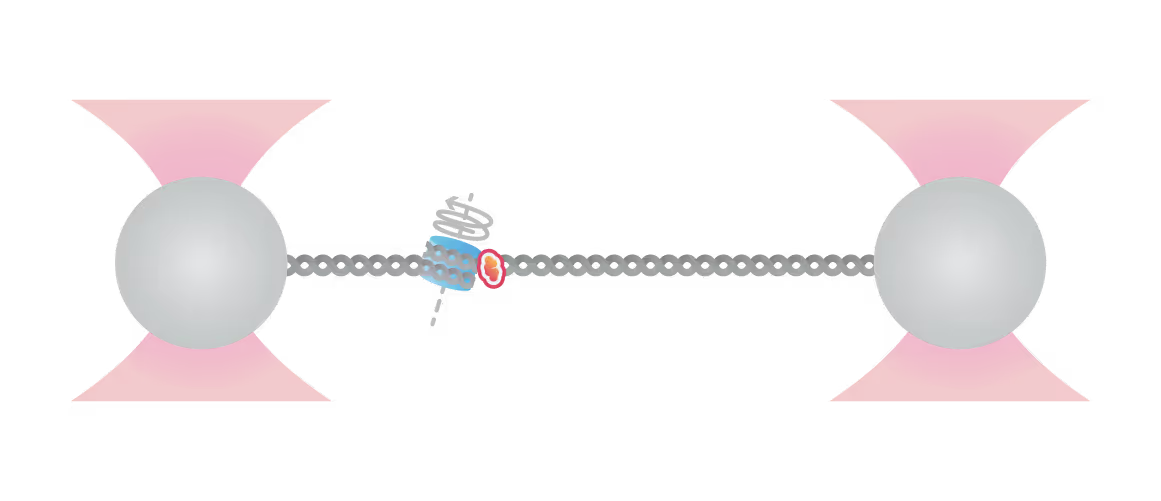
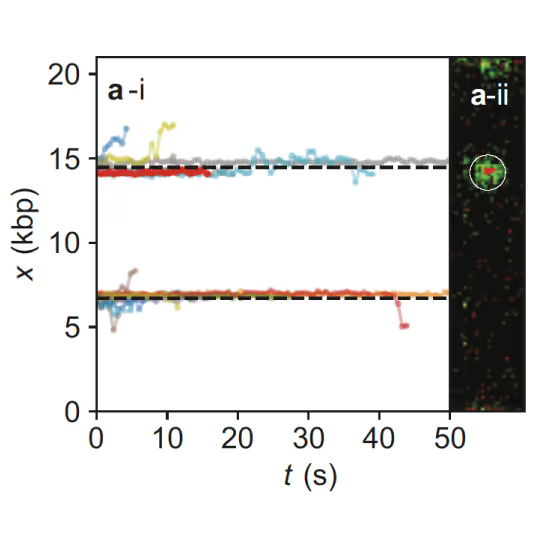
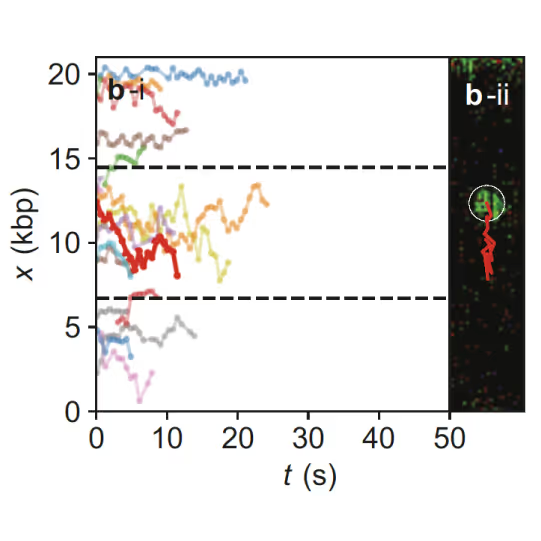
Samples of tracking data (i) and confocal images (ii) illustrating dynamics of ORC initially bound within the origin sequence (a) or at a non-specific location (b). Figure from Sánchez, H., McCluskey, K., van Laar, T. et al. DNA replication origins retain mobile licensing proteins. Nat Commun 12, 1908 (2021). https://doi.org/10.1038/s41467-021-22216-x
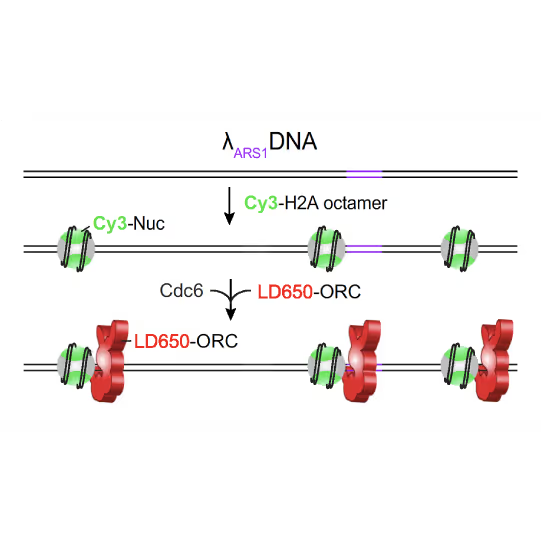
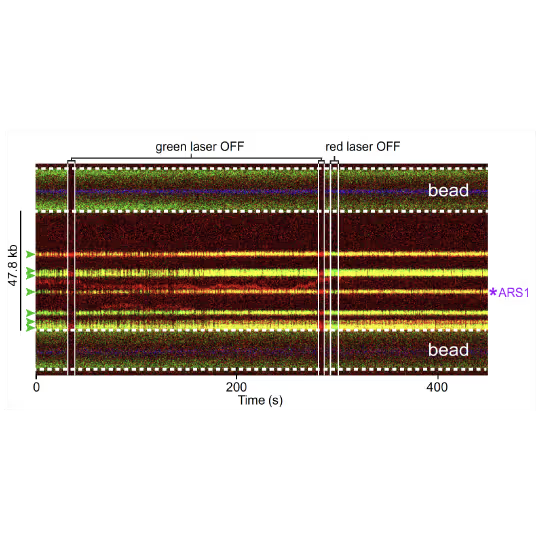
(a) Cartoon DNA containing an ARS sequence sparsely loaded with nucleosomes (green) and incubated with ORC (red) and Cdc6. (b) A representative kymograph showing a λARS1 DNA tether loaded with multiple nucleosomes (positions indicated by green arrowheads), all of which were located at non-ARS1 sites except one. Each nucleosome was observed to be stably bound by ORC (red). Figure from Li, S., Wasserman, M.R., Yurieva, O. et al. Nucleosome-directed replication origin licensing independent of a consensus DNA sequence. Nat Commun 13, 4947 (2022). https://doi.org/10.1038/s41467-022-32657-7
Dive into the publication

Studying DNA and chromatin replication using integrated force-fluorescence microscopy
Chromatin replication is a highly complex process and is crucial for genome integrity, and thus the proper functioning and survival of any organism. Copying chromatinized DNA with its sophisticated structural elements like nucleosomes and structural maintenance of chromatin (SMC) protein-induced loops as well as various modifications and possible damage sites poses quite a challenge to the molecular replication system (replisome).
In this webinar, Prof. Nynke Dekker explains how she and her team have used Dynamic Single-Molecule (DSM) microscopy to identify and quantify fundamental molecular interactions between the origin recognition complex (ORC) and minichromosome maintenance protein (MCM) complex in the context of nucleosomes.
Replication in context: Uncover mechanisms ensuring replication fidelity and genome stability
In cells, DNA replication occurs in a complex environment of DNA structures and in parallel to processes like transcription and DNA repair. Moreover, correction steps ensuring the proper reproduction of the genome need to be carried out throughout the replication process. Direct insights into the underlying molecular mechanisms are key to our understanding of replication as an integrated cellular process.
Various authors revealed key information on replication mechanisms:
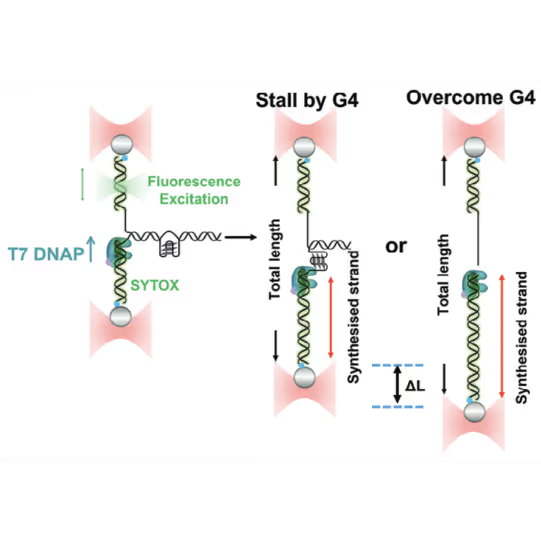
- In the lab of Bo Sun, direct observation of replisomes encountering G4 quadruplexes shed light on how replication collapse is avoided by complex interplay of helicases and polymerases resolving the quadruplex structure
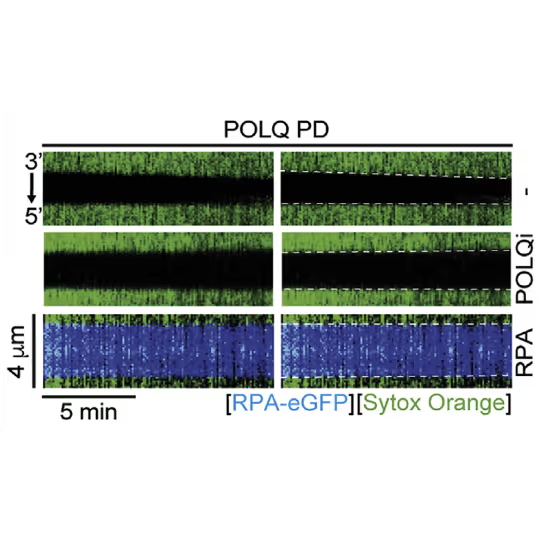
- The labs of Stephen West, Simon Boulton and David Rueda identified the role of POLQ in repairing post-replicative ssDNA gaps which opens new approaches to treating BRCA deficient cancers
Further reading
Belan et al. Molecular Cell (2022)
Guo et al. Advanced Science (2023)
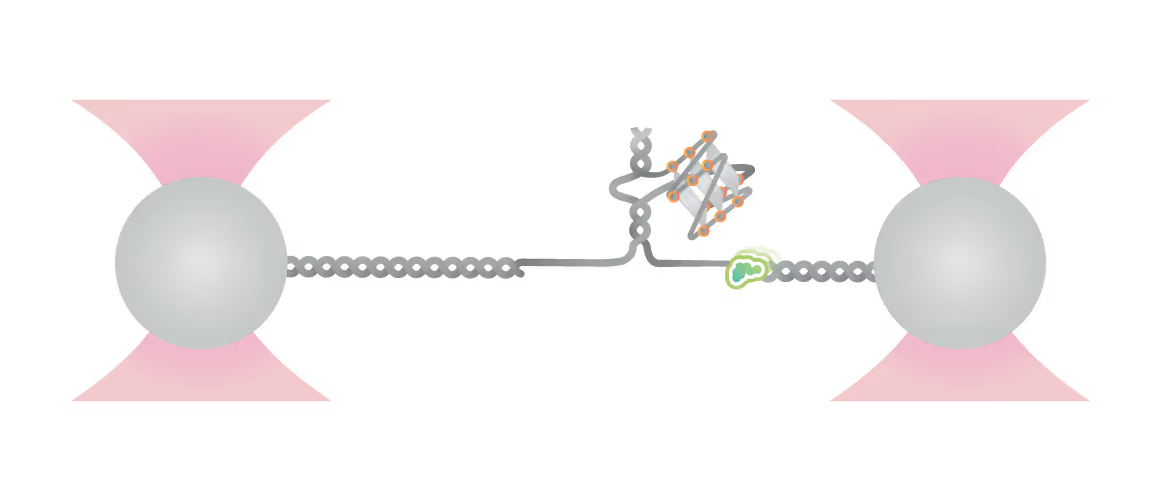
Schematic diagram of possible outcomes of T7 DNAP synthesis encountering a leading-strand G4 in the optical tweezer assay. A T-shaped DNA template harboring a leading-strand G4 is suspended between two optical traps while DNAP activity is monitored with fluorescence imaging. Figure from Guo, L. et al. Joint Efforts of Replicative Helicase and SSB Ensure Inherent Replicative Tolerance of G‐Quadruplex. Advanced Science 2307696 (2023). doi:10.1002/advs.202307696
Kymographs showing the extension of 3ʹ DNA end at the edge of the ssDNA gap by POLQ PD. dsDNA (green) in the presence or absence of 1 nM RPA-EGFP or 10 μM POLQ inhibitor (POLQi). When present, RPA signal is shown in blue indicating ssDNA. DNA extension rate was measured as a slope of the border of spreading dsDNA signal. Figure from Belan et al., POLQ seals post-replicative ssDNA gaps to maintain genome stability in BRCA-deficient cancer cells, Molecular Cell (2022), https://doi.org/10.1016/j.molcel.2022.11.008
Dive into the publication
C-Trap
Molecular biology as never seen before
The C-Trap® provides the world’s first dynamic single-molecule microscope to allow simultaneous manipulation and visualization of single-molecule interactions in real time.

These cards are NOT components because they use the finsweet nested collection logic. To pull in the posible multiple people wo worked on it.
This type of 1-many relation is not supported native in Webflow.
Also this section is hidden when emtpy. To keep everything visible here, that is being done outside the webflow designer from within Slater.
DNA polymerase actively and sequentially displaces single-stranded DNA-binding proteins
Mapping fast DNA polymerase exchange during replication
Replication Fork Activation Is Enabled by a Single-Stranded DNA Gate in CMG Helicase
SITC 2025
CAR-TCR Summit 2025
CICON 2025