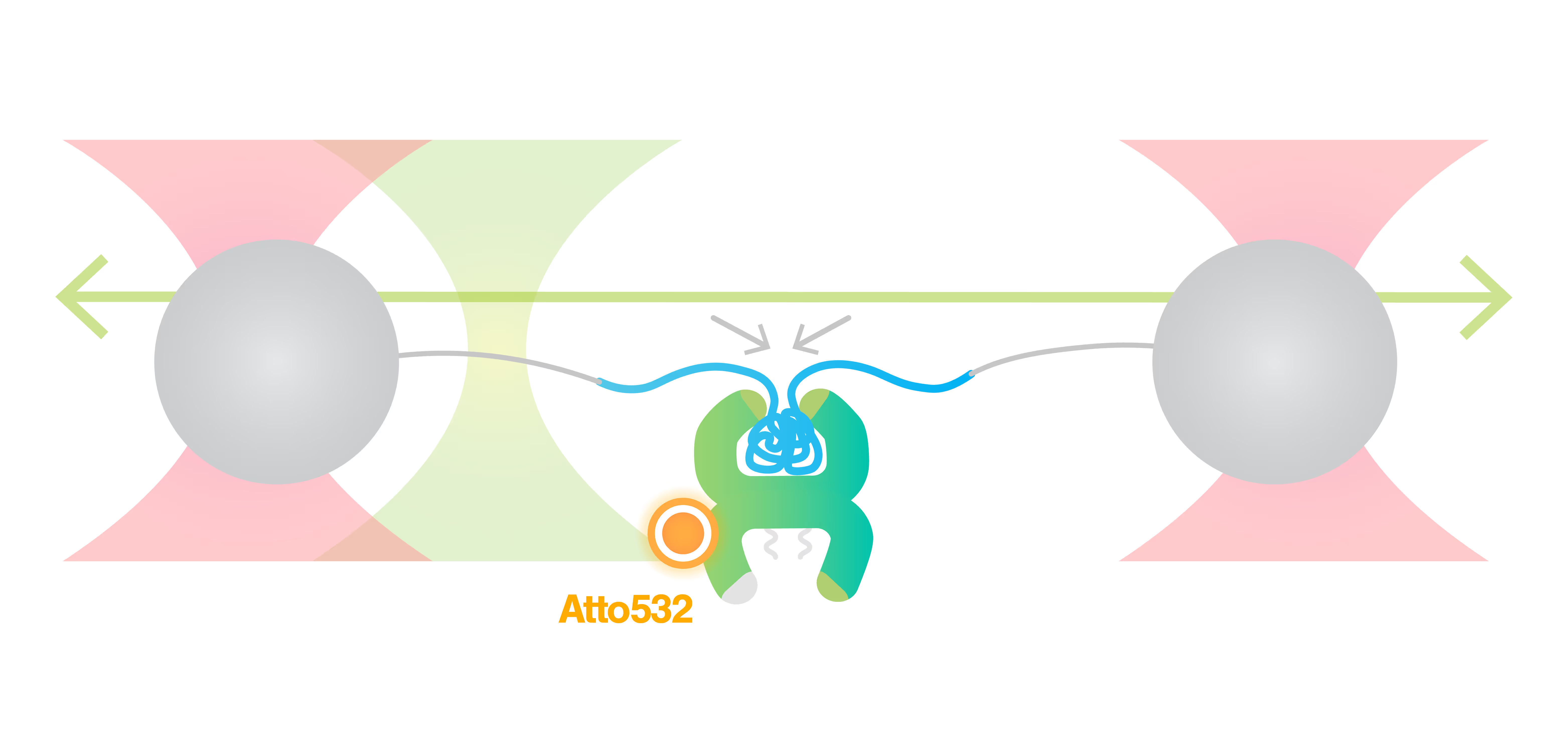
A new view on the structure-function relationship of proteins
Study of multi-domain protein folding & conformational dynamics
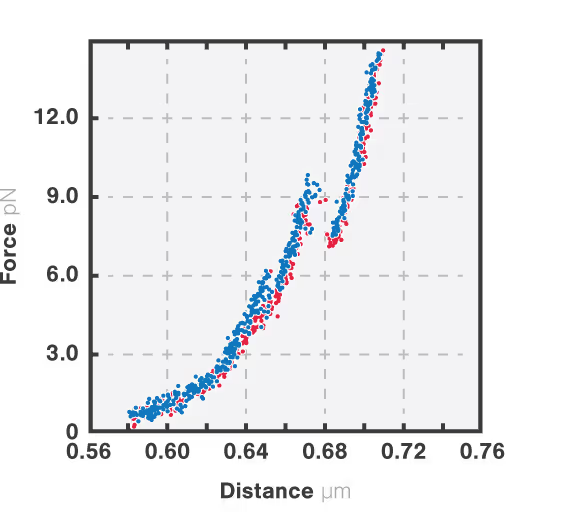
The resulting force-distance curve (Figure 1) of calmodulin – a calcium-binding protein – reveals that the protein unfolds and refolds in two steps which correspond to two separate helix-loop-helix domains. By simultaneously observing the FRET fluorescence signal, it can be seen how the unfolding of the domains results in fluctuations of the FRET signal due to changes in the relative position of the FRET pair.
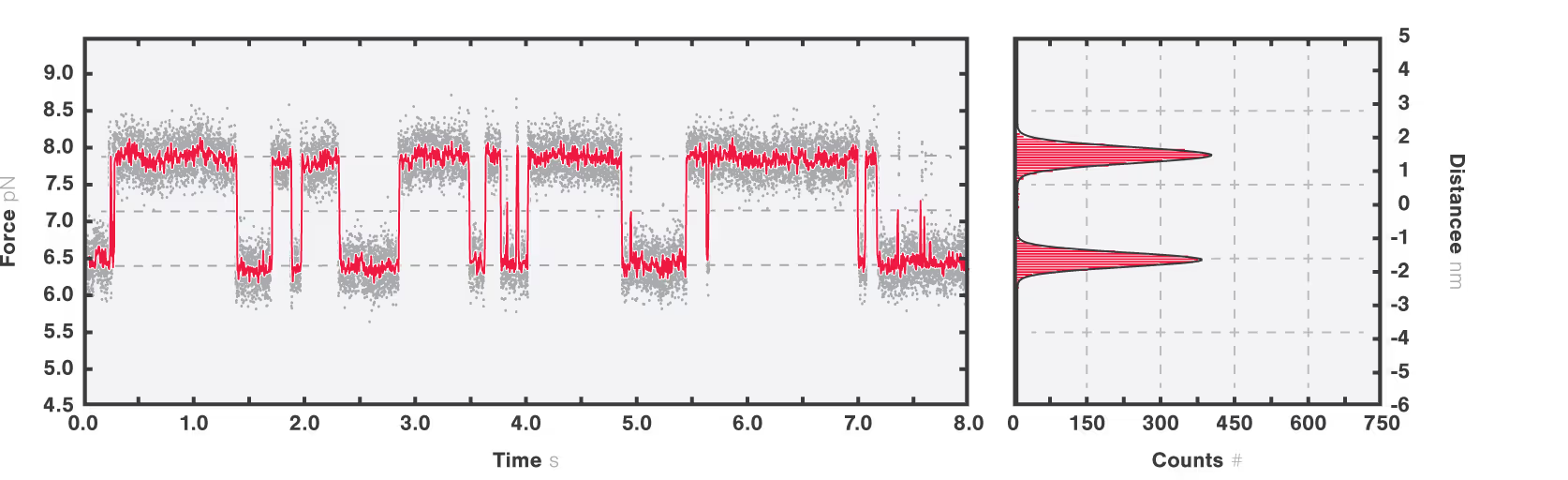
Equilibrium dynamics showing the transition between short-lived intermediate states can be studied because of the intrinsic distance clamp which keeps the traps at a fixed distance, while force fluctuations are measured. When this is applied to calmodulin, the equilibrium fluctuations between the states can be observed with a force resolution of <0.1 pN at 100 Hz. Figure 2 shows that calmodulin switches between two major states, without a clear preference and that intermediate steps can be resolved as calmodulin occasionally jumps to a third state for short periods of time, represented by the dashed grey lines.
Sample courtesy of Prof. Carlos Bustamante at the University of California, Berkeley

Figure 2: Full length CaM protein at 10 mM Ca2+ showing equilibrium dynamics between multiple states. Data is recorded at 50 kHz (grey line) and averaged at 200 Hz (red line). The histogram quantifies the most populated states in the inset (right panel) showing two peaks at 6.5 ± 0.1 pN and 7.8 ± 0.09 pN (mean ± standard deviation).
Dive into the publication
Explore further

Protein folding and conformational changes
The biological function of macromolecules such as proteins and RNA is intimately linked to their conformation and dynamic structural changes. Proper folding into native conformations is critical for function, while misfolding can lead to loss of activity or the onset of diseases, including neurodegenerative disorders associated with protein aggregation.
Understanding the mechanisms of protein folding and conformational transitions is therefore essential for elucidating biological processes and disease pathways. Single-Molecule Force Spectroscopy (SMFS) offers a powerful approach to probe these mechanisms by isolating individual molecules and monitoring real-time conformational changes under applied mechanical force. This application note presents a demonstrative experiment utilizing high-resolution optical tweezers developed by LUMICKS to investigate the folding pathway of Calmodulin (CaM), the primary calcium-binding protein in the human body.
The results highlight the capability of SMFS to provide high-sensitivity measurements of protein conformational dynamics at the sub-nanometer scale.
C-Trap
Molecular biology as never seen before
The C-Trap® provides the world’s first dynamic single-molecule microscope to allow simultaneous manipulation and visualization of single-molecule interactions in real time.

These cards are NOT components because they use the finsweet nested collection logic. To pull in the posible multiple people wo worked on it.
This type of 1-many relation is not supported native in Webflow.
Also this section is hidden when emtpy. To keep everything visible here, that is being done outside the webflow designer from within Slater.
One-pot dual protein labeling for simultaneous mechanical and fluorescent readouts in optical tweezers
Co-translational ribosome pairing enables native assembly of misfolding-prone subunits
Single-Molecule Insight Into α-Synuclein Fibril Structureand Mechanics Modulated by Chemical Compounds
SITC 2025
CAR-TCR Summit 2025
CICON 2025