The precision and dynamics behind genome engineering
Understand off-target activities of Cas for safer gene editing
Current CRISPR/Cas research is focused on fully understanding Cas function under different circumstances to exclude off-target binding events and ensure its precision during DNA editing applications. With experiment and data analysis automation, the C-Trap enables a fast workflow for precise localization of DNA-binding proteins. Critical parameters like the location and probability of off-target binding and activity under different conditions become readily accessible.
Dynamic single-molecule research by the groups of David Rueda(MRC-LMS) and Guillermo Montoya (University of Copenhagen) revealed that:
- Moderate tension on DNA induces Cas9 and Cas12a off-target binding
- Off-target binding sites relate to the local stability of the DNA double-strand and thus its sequence
- Off-target dsDNA-ssDNA intermediates rather than stable ssDNA attract Cas
- Supercoiling has a similar effect as tension and increases unspecific interactions
Taken together, these findings improve our understanding of CRISPR/Cas gene editing mechanisms and open up new approaches of increasing gene editing reliability and safety.
Further reading:
- Newton et al. Nat Struct Mol Biol (2019)
- Losito et al Phys. Chem. Chem. Phys. (2021)
- Paul et al. Nucleic Acids Research (2021)
- Newton et al. Molecular Cell (2023)

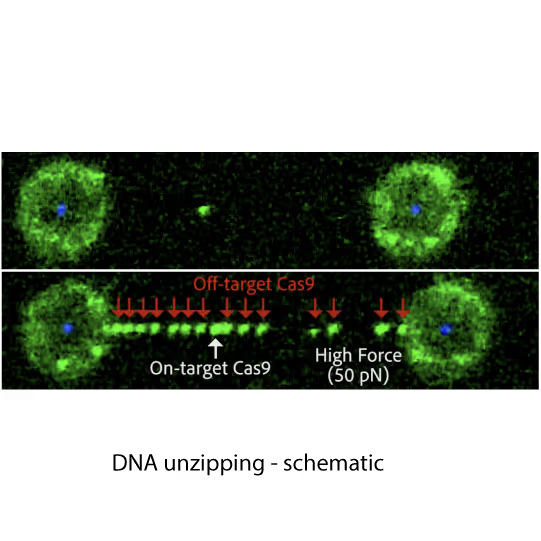
Fluorescence images showing dCas9 on-target binding to DNA at contour length (top) and additional off-target binding (bottom) to stretched DNA that has formed bubbles after applying 20 pN force. Left and right fluorescent spheres are optically trapped polystyrene beads that are holding the DNA molecule (unlabelled) in between.
Dive into the publication

Unlocking the power of Cas12a: Novel insights into engineered endonucleases for safe and precise next generation genome editing
The advent of powerful and precise gene-editing tools is transforming the way we approach health, medicine, and biological research, opening up possibilities that were once considered science fiction.
Join us today in this webinar to unravel the secret of Cas12a endonuclease, a component of the revolutionary CRISPR-Cas gene-editing technology. Prof. Guillermo Montoya will guide us through his groundbreaking research that explores how Cas12a, an RNA-guided enzyme, interacts with bacteriophage λ-DNA. This journey into the world of genome editing will not only deepen our understanding of this complex tool but also inspire us to imagine the potential advancements in biomedicine and biotechnology.
As we embark on this journey together, we are excited to share and explore the novel insights and possibilities that are becoming accessible through these advancements in gene-editing technologies. Let’s dive in and explore the extraordinary world of Cas12a and its implications in gene editing.
Explore further

Unlock the true potential of CRISPR Cas9 technology through real-time direct visualization of Cas9 gene-editing
CRISPR Cas9 is a gene editing tool that has increased in popularity due to its simplicity to use. It allows researchers to seamlessly edit DNA sequences by combining a sequence identifying guide RNA (gRNA) with the Cas9 endonuclease enzyme. However, its applicability as a gene-editing and therapeutic tool is impeded due to undesired off-target binding of Cas9.
Conventional bulk in vitro assays provide information on the DNA sequences but do not give insights into the changes in the local DNA structure. In this application note, dynamic single molecule (DSM) analysis is shown to be a novel and a more powerful investigative tool to directly visualize and manipulate CRISPR Cas9 – DNA interactions, and accelerate gene-editing researchand therapy development.
Real-time insights into gene editing mechanisms
Understanding the molecular interactions that govern Cas-DNA binding is crucial for the development of safe and reliable medical applications of the CRISPR/Cas gene editing system. Recent research indicates that sequence-matching interactions between the guide RNA and DNA template are not the only drivers of Cas binding and catalytic activity.
Bo Sun and his group at Shanghai Tech utilize Dynamic Single-Molecule microscopy with the C-Trap to take a closer look at the interactions of Cas9 and its substrate. They were able to identify and characterize stable interactions between different Cas9 variants and DNA beyond the protospacer adjacent motif (PAM), leading to valuable insights:
- Cas9 interacts with DNA sequences up to 14 basepairs downstream of the PAM
- This interaction is critical for stable DNAbinding and cleavage activity of Cas9
- Upon DNA cleavage, Cas9 releases the PAM-distalDNA, thus facilitating its dissociation
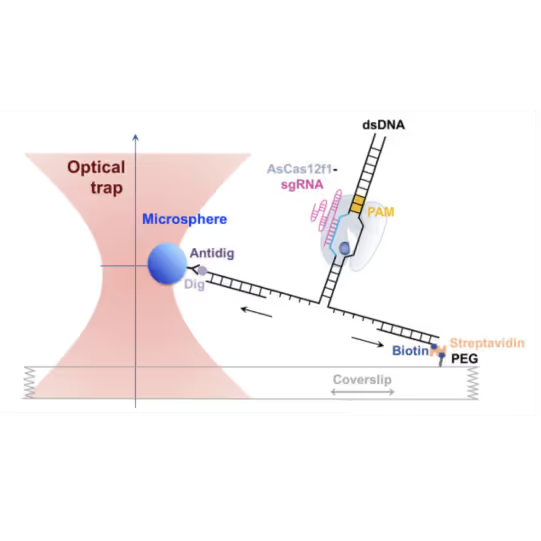
- Unzipping DNA with bound Cas9 variants reveals that the stable interaction is mediated by electrostatic interactions between specific amino acids and the DNA backbone
- AsCas12f1, utilized as a versatile gene delivery platform, combines out-of-protospacer DNA unwinding with exonuclease activity in the sequential target cleavage
Dive into the publication
C-Trap
Molecular biology as never seen before
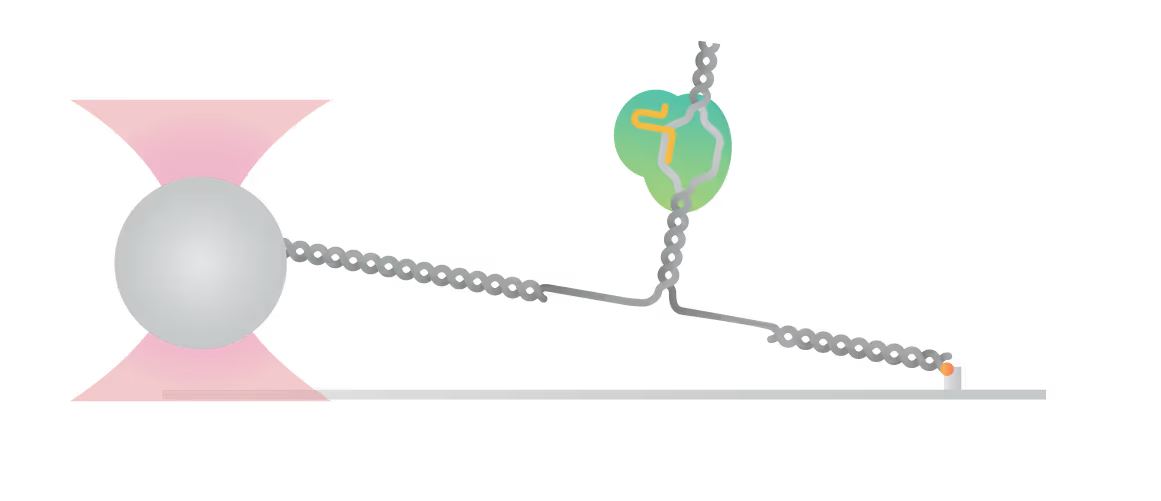
The C-Trap® provides the world’s first dynamic single-molecule microscope to allow simultaneous manipulation and visualization of single-molecule interactions in real time.

These cards are NOT components because they use the finsweet nested collection logic. To pull in the posible multiple people wo worked on it.
This type of 1-many relation is not supported native in Webflow.
Also this section is hidden when emtpy. To keep everything visible here, that is being done outside the webflow designer from within Slater.
The post-PAM interaction of RNA-guided spCas9 with DNA dictates its target binding and dissociation
DNA stretching induces Cas9 off-target activity
Dynamics of Staphylococcus aureus Cas9 in DNA target Association and Dissociation
SITC 2025
CAR-TCR Summit 2025
CICON 2025