The forces and dynamics behind cellular architecture and transport
Investigation of motor protein stepping mechanics along cytoskeletal filaments
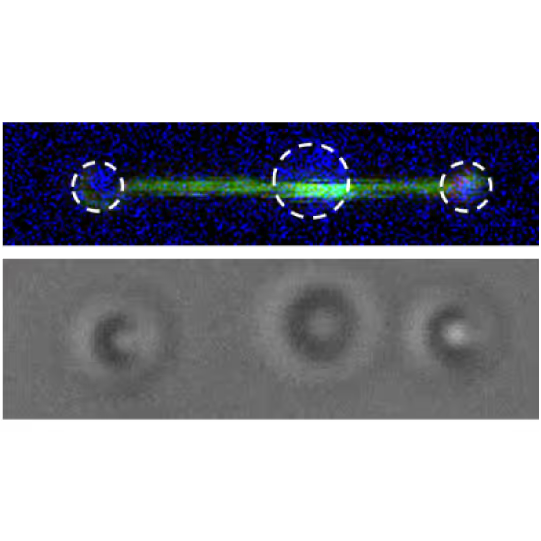
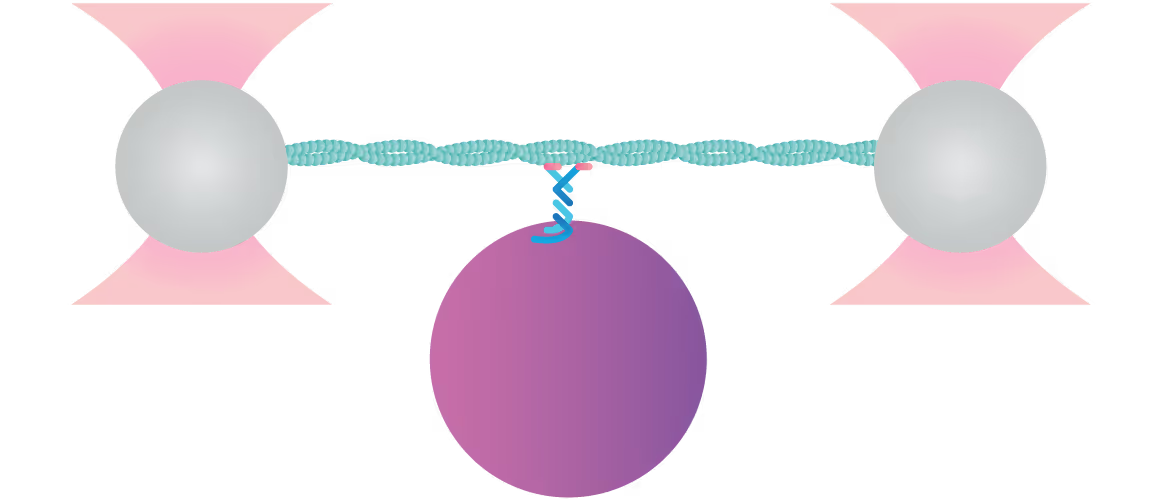
Here, a three-bead assay is utilized to study the stepping behavior of a molecular motor. Two optically-trapped beads hold actin tight and a third myosin-coated bead is immobilized on the glass surface of a flow cell. We can visualize the fluorescently-tagged actin filaments using correlated confocal microscopy and perform simultaneously distance measurements. This offers the possibility to track the activity of molecular motors along cytoskeletal proteins and determine their thermodynamic properties at the single molecule level.
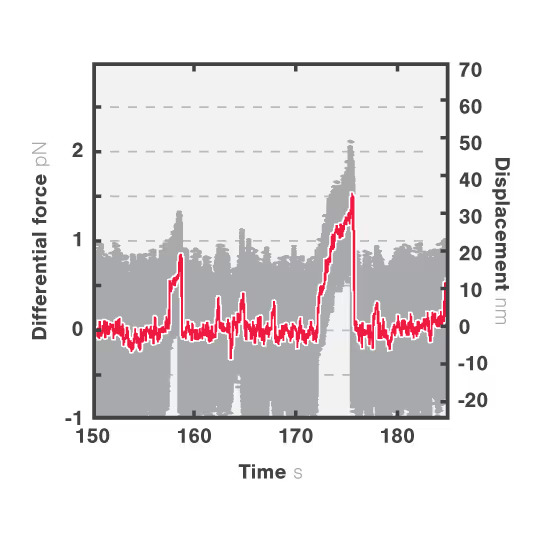
In this assay, we investigated the displacement of actin with respect to wild-type myosin-VI by measuring the correlated displacements of the two beads trapped in the dual optical trap. Figure 1 shows that myosin pulls the actin filament in a unidirectional manner with a measurable force.
The exact kinetics of the motor can be observed by measuring the relative translocation of the filament with respect to the motor and the related forces. From the resulting force–time traces, we can derive the speed and processivity of the motor.
Sample courtesy of Prof. Dr. Margaret Titus at the University of Minnesota
Dive into the publication
Stepping of filaments and motors at the surface
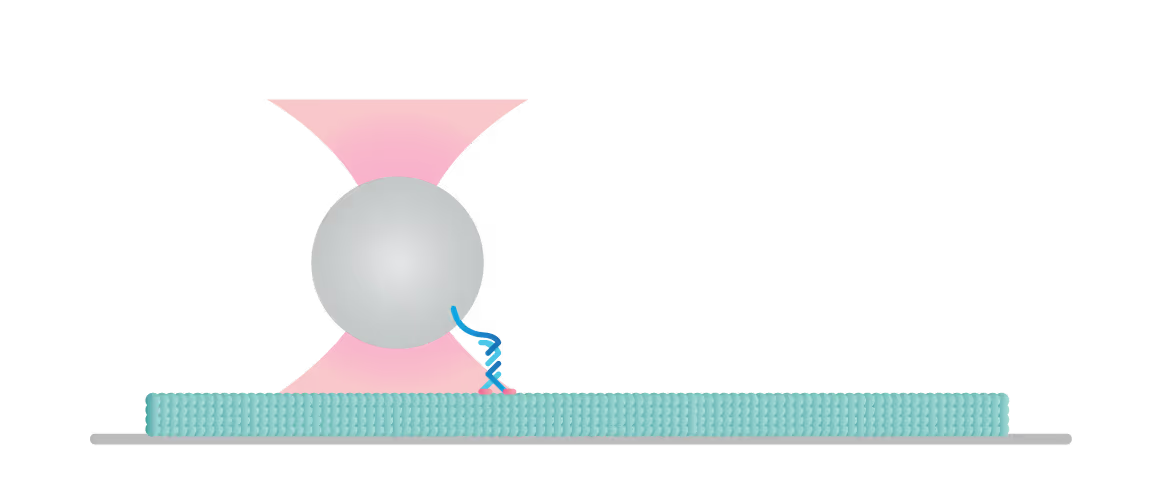
The C-Trap enables you to construct and test a variety of assays to understand the molecular mechanisms of motor-filament interactions and gain insights into their regulation. In this experiment we tethered a kinesin motor to a bead and placed the motor on a microtubule which we visualized with label-free IRM.
We can follow the motor’s activity in all 3 dimensions as it moves over the filament and measure the distance, velocity, and (stall-) force of a motor attached to a bead, also under the influence of other molecules and loads.
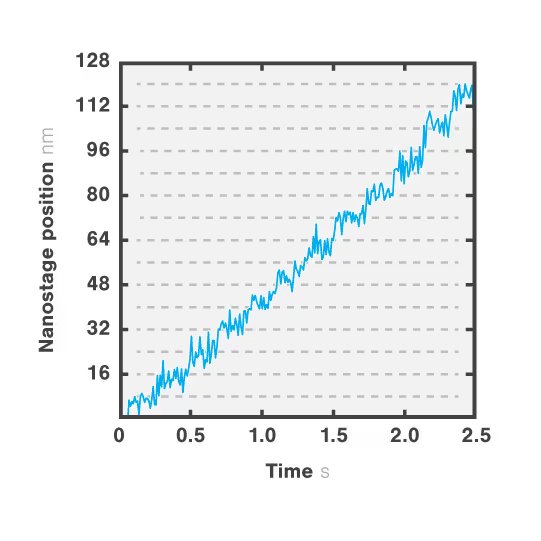
Figure 2 shows a single kinesin motor stepping along a microtubule. Steps, 8 nm in size, were collected with 2D feedback along the microtubule direction at stall force of 1 pN.
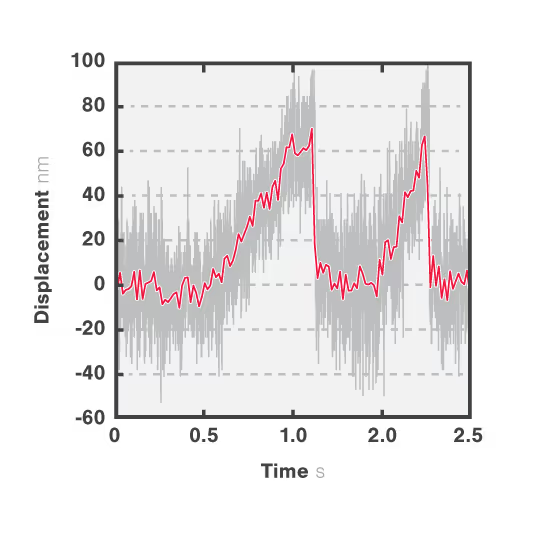
In Figure 3 we can observe 2 events during which the kinesin motor detaches from the microtubule at a force of approximately 2 pN.
Data courtesy of Dr. Paul Ruijgrok and Dr. Zev Bryant at Stanford University.
Dive into the publication
Force-extension, manipulation, and visualization of polymers and protein filaments
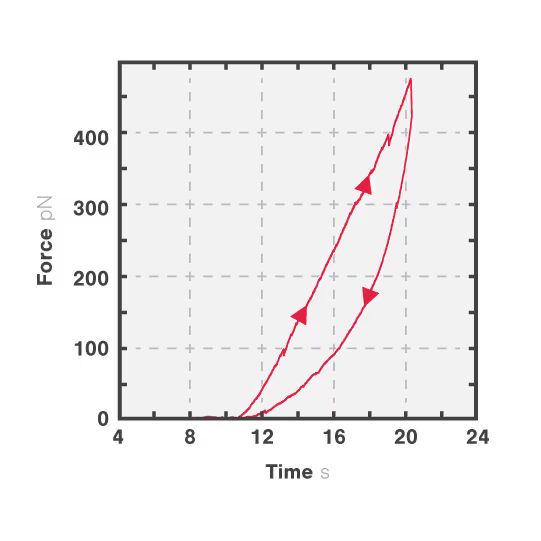
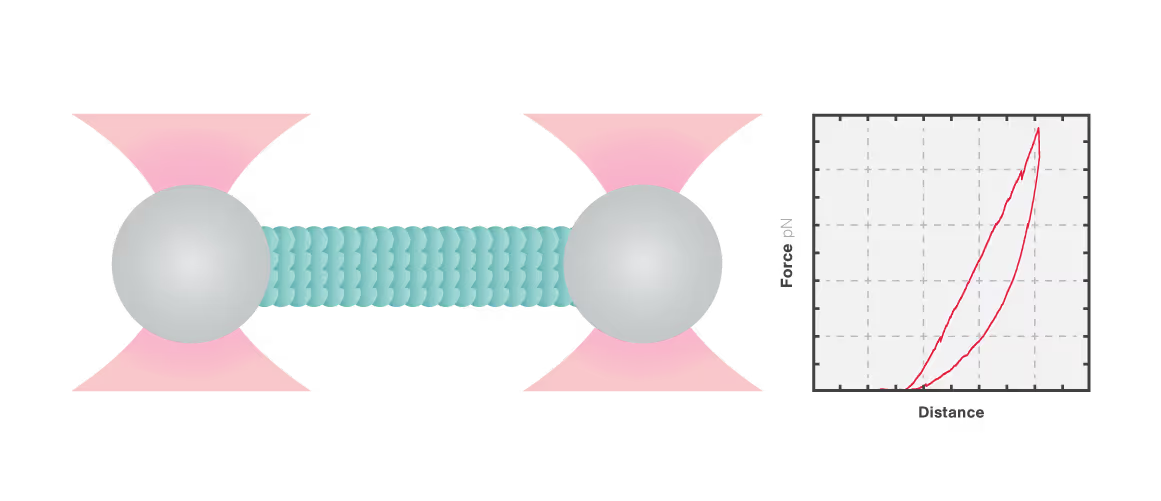
In this experiment, a vimentin filament is tethered between two optically trapped beads. We can obtain the force-distance curve and determine the mechanical properties of the microtubule by simultaneously stretching the filament and measuring the force and the extension.
The measured data in Figures 1 and 2 show the extension of a vimentin intermediate filament. The force-distance curve is measured while stretching and relaxing the intermediate filament at a slow speed, allowing for structural equilibrium. The retraction curve shows clear hysteresis due to the remodeling of the vimentin filament under high tension. Simultaneous confocal fluorescence imaging of the vimentin filament is used to resolve this intra-molecular remodeling.
Combining optical tweezers with simultaneous fluorescence measurements allows correlating the mechanical properties of the microtubule with local information.
Dive into the publication
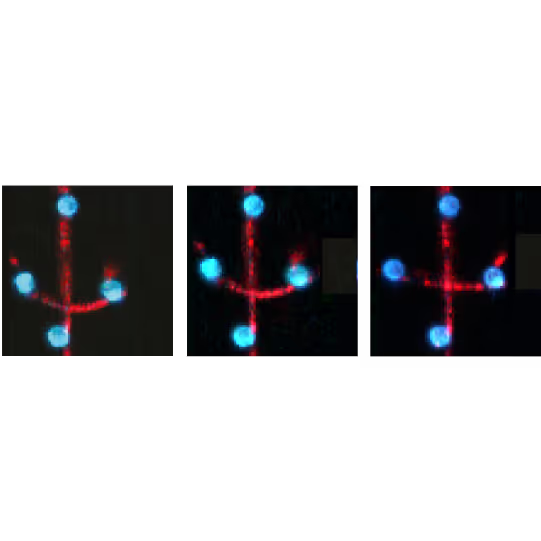
Visualization of filament-filament interactions
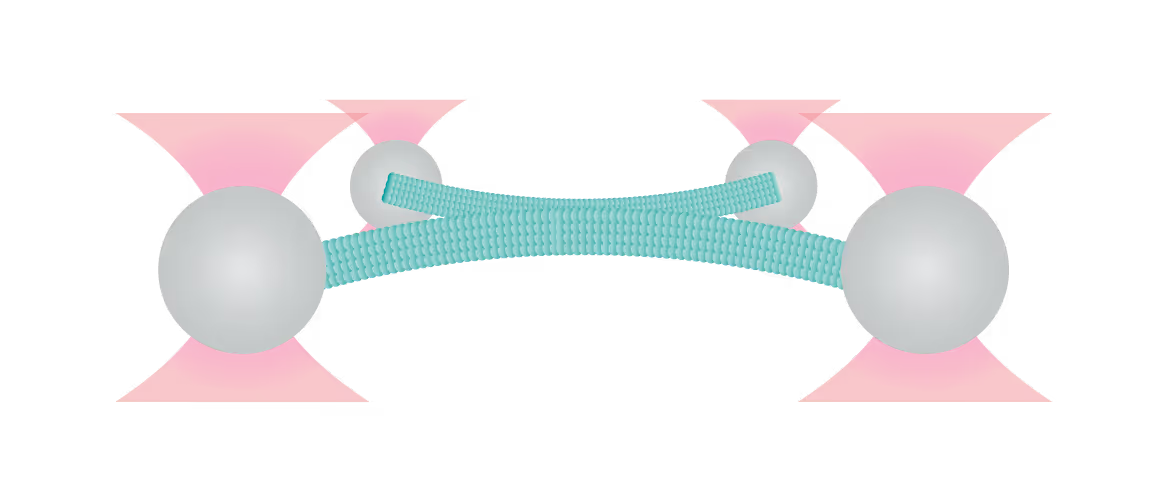
Figure 1 shows the interaction between two fluorescently labeled microtubules, as one microtubule is dragged across the other with a known force. In the middle and left snapshot, we can observe that the friction force between the filaments causes one microtubule to adopt a curved conformational structure. When we stop the movement of the microspheres the microtubule slowly relaxes (right).
The surface properties of these filaments, the electrostatic and ionic forces and specific interactions, can generate friction when two filaments interact. This friction can be measured using a device such as the C-Trap® configured for quadruple optical trapping.
Dive into the publication
C-Trap
Biomolecular interactions re-imagined
The C-Trap® provides the world’s first dynamic single-molecule microscope to allow simultaneous manipulation and visualization of single-molecule interactions in real time.

These cards are NOT components because they use the finsweet nested collection logic. To pull in the posible multiple people wo worked on it.
This type of 1-many relation is not supported native in Webflow.
Also this section is hidden when emtpy. To keep everything visible here, that is being done outside the webflow designer from within Slater.
Supported Natural Membranes on Microspheres for Protein–Protein Interaction Studies
Tetraspanin 4 stabilizes membrane swellings and facilitates their maturation into migrasomes
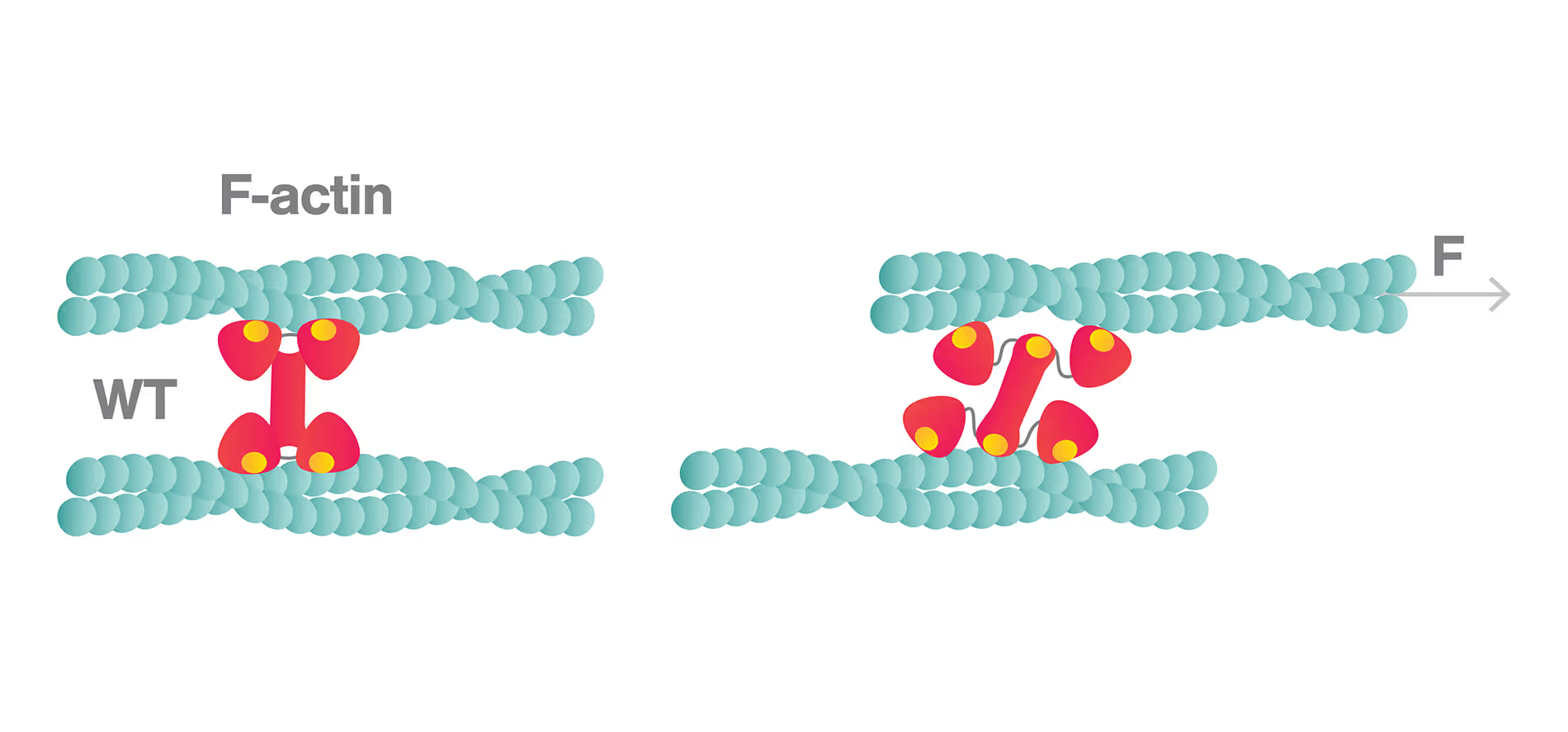
Weak catch bonds make strong networks
SITC 2025
CAR-TCR Summit 2025
CICON 2025