The complexity of transcription mechanisms
- Visualize single DNA polymerase in real-time
- Base-pair level observation of DNA transcription dynamics
- Observe while manipulate the transcription process at the same time
Real-time observation of DNA exonuclease dynamics at base-pair level
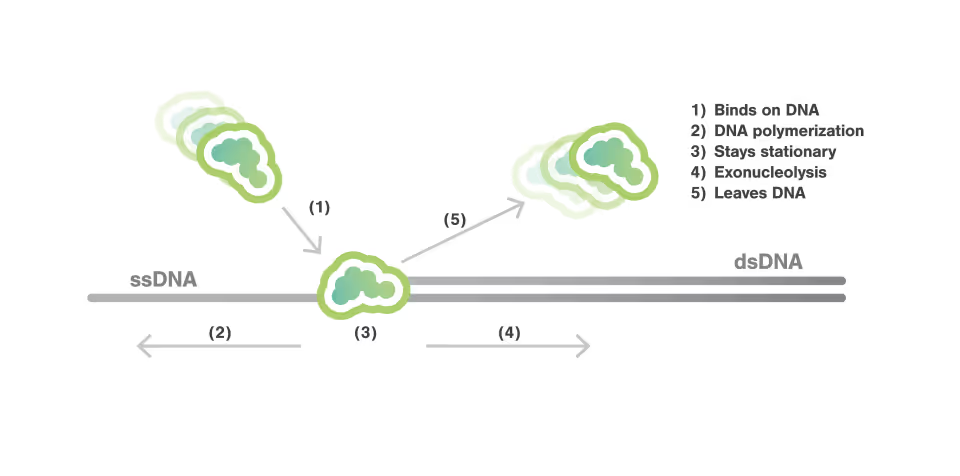
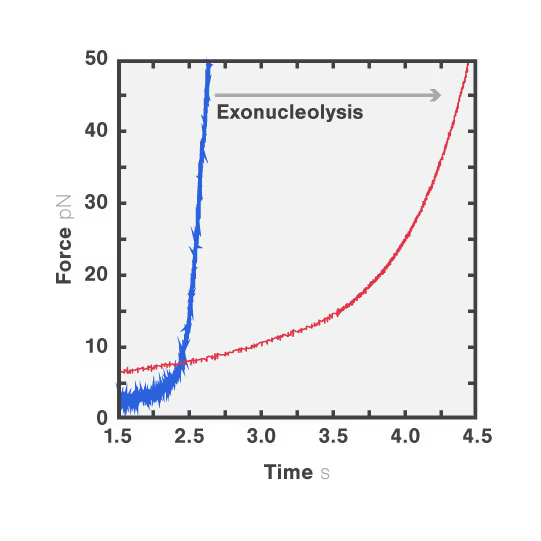
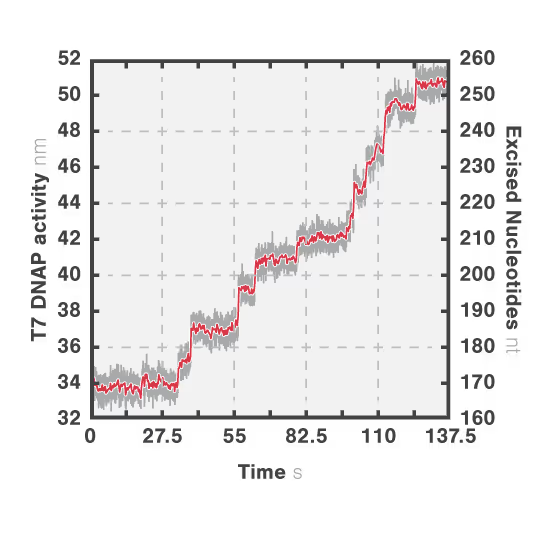
In this experiment, the exonuclease activity of the T7 polymerase was investigated. This is a DNA polymerase from the T7 bacteriophage that copies DNA strands in the 5’→ 3′ direction, and also features exonuclease activity. For this analysis, an optically trapped double-stranded DNA was held at a constant force that induced exonucleolytic activity of the polymerase. By removing nucleotide after nucleotide from one strand, the polymerase was unwinding the dsDNA. As the length of single-stranded DNA is longer compared to its double-stranded state, the unwinding resulted in a gradual increase in the end-to-end distance of the DNA (Figure a). This change in length was directly translated into the activity of the T7 DNA polymerase and the number of nucleotides it processed over time (Figure b). Specifically, short activity bursts ranging between 3 and 10 nucleotides were revealed, interspersed by frequent pauses of varying duration. This provided deeper insights into the dynamics of T7 exonuclease activity.
Dive into the publication
C-Trap
Molecular biology as never seen before
The C-Trap® provides the world’s first dynamic single-molecule microscope to allow simultaneous manipulation and visualization of single-molecule interactions in real time.

These cards are NOT components because they use the finsweet nested collection logic. To pull in the posible multiple people wo worked on it.
This type of 1-many relation is not supported native in Webflow.
Also this section is hidden when emtpy. To keep everything visible here, that is being done outside the webflow designer from within Slater.
Kinetic control of mammalian transcription elongation
Phase-separated NDF−FACT condensates facilitate transcription elongation on chromatin
Chromatin sequesters pioneer transcription factor Sox2 from exerting force on DNA
SITC 2025
CAR-TCR Summit 2025
CICON 2025


.avif)