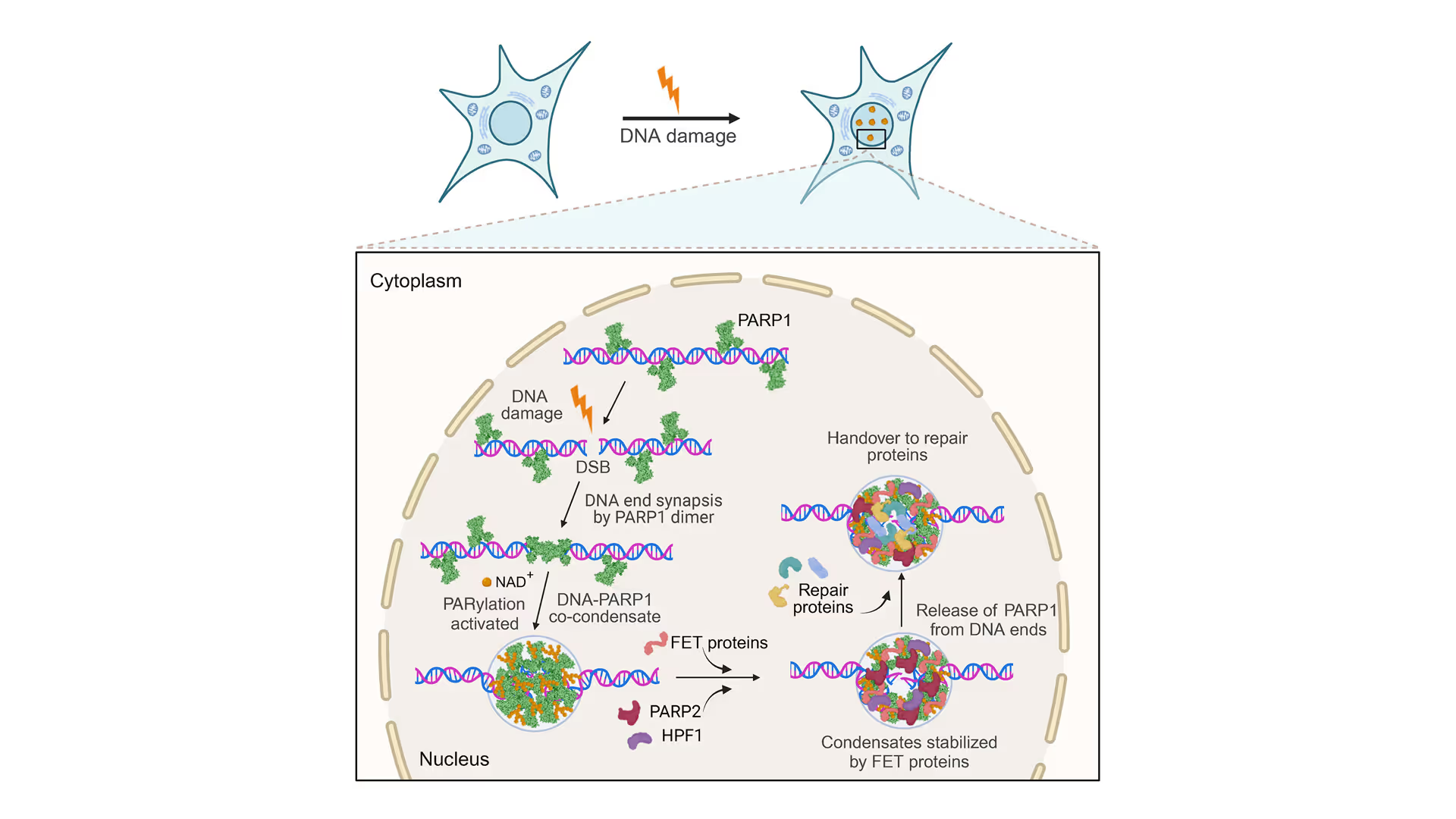
DNA double-strand breaks (DSBs) are critical lesions that, if not properly repaired, can result in chromosomal rearrangements and genomic instability, underlining the importance of efficient repair systems in maintaining cellular and genetic health. The cell’s response involves a range of complex repair mechanisms such as homologous recombination and non-homologous end joining that are critical for the prevention of DSB-induced cell death, mutations, and cancer.
PARP1(poly[ADP-ribose] polymerase 1) plays a crucial role in DNA damage repair by detecting DNA strand breaks and facilitating the repair process. It acts as a signaling molecule that recruits DNA repair proteins to the site of damage and is involved in several DNA repair pathways. PARP1 inhibitors have been effectively utilized in cancer treatment, offering a targeted approach to disrupt the DNA repair processes in tumor cells. Despite its established role, the detailed process of how DSB sites assemble and how PARP1 prevents the separation of broken DNA strands remains poorly understood.
Common DNA repair pathways
- Nucleotide excision repair (NER) detects and repairs bulky DNA lesions, such as those caused by UV light, by removing the damaged section and synthesizing new DNA to replace it
- Base excision repair (BER) is a cellular mechanism that corrects DNA containing small, non-helix-distorting base lesions, ensuring genomic integrity by excising and replacing damaged bases.
- Non-homologous end joining (NHEJ) is a repair mechanism for DNA double-strand breaks that directly ligates the broken ends together without needing a homologous template, which can sometimes lead to mutations.
- Microhomology-mediated end joining (MMEJ) repairs DNA breaks by aligning short, homologous sequences near the break points, often resulting in deletions due to the loss of intervening DNA
- Homologous recombinational repair (HRR) fixes DNA double-strand breaks using a homologous sequence as a template, ensuring accurate repair and maintaining genetic integrity
- DNA mismatch repair (MMR) identifies and corrects mismatched nucleotides (errors introduced during DNA replication) to prevent mutations, thereby maintaining the DNA’s genetic accuracy.
Extensive research has delved into the role of PARP1 in DNA repair. Notably, Prof. Ben van Houten’s lab utilized single-molecule analysis to uncover how tension impacts PARP1’s ability to bind to DNA nicks. For a deeper dive into these findings, you can watch Prof. Ben van Houten’s on-demand webinar.
In a recent Cell study led by Simon Alberti’s group from the Dresden University of Technology, it was shown that PARP1 plays a vital role in DNA repair by preventing the separation of broken DNA ends through a groundbreaking novel mechanism involving co-condensation with DNA. Employing LUMICKS C-Trap technology, the team was able to demonstrate that PARP1-DNA condensates are key to securing broken DNA ends which was not possible with other technologies. This was achieved by precisely manipulating single DNA molecules and measuring their tension and length, providing concrete evidence of the condensates’ essential function in maintaining DNA strand integrity following damage.

Single-molecule analysis of cancer DNA-protein interactions from nuclear extracts
Understanding of DNA repair mechanisms could advance treatments for cancer and diseases of aging. But reconstituting DNA repair protein complexes from cancerous tissues to study their mechanisms of action is often time-consuming or, in some cases, impossible. A new technique performing dynamic single-molecule analysis directly on nuclear extracts allows rapid mechanistic analysis of mutant proteins from cancer cells, providing previously unseen insights into their mechanisms of action. This new innovative tool, when combined with rapid data analysis, represents a bridge between the study of biochemistry of purified proteins and molecular biology.
What are biomolecular condensates?
In cell biology, condensates refer to membrane-less organelles or structures formed through phase separation, where specific proteins and nucleic acids aggregate to create distinct compartments within the cell without being enclosed by a membrane. Condensates have the unique ability to locally and reversibly enrich enzymes, substrates, and various molecules, playing a pivotal role in cellular processes. Research in the past ten years highlights their crucial roles in streamlining and regulating cellular activities.
Using fluorescence imaging, the authors first demonstrated that PARP1 assembles with dsDNA free ends to form condensates. These condensates’ enzymatic activity is fine-tuned through PARylation, alongside the regulation by various repair proteins, showcasing the control mechanisms that govern DNA repair processes.
Following this, C-Trap technology for the first time provided insights into how these PARP1-DNA condensates physically secure the DNA’s free ends. This goes beyond simply gathering components; it establishes a stable environment essential for effective repair, ensuring the DNA strand’s integrity and functionality throughout the process.
How were the experiments conducted?
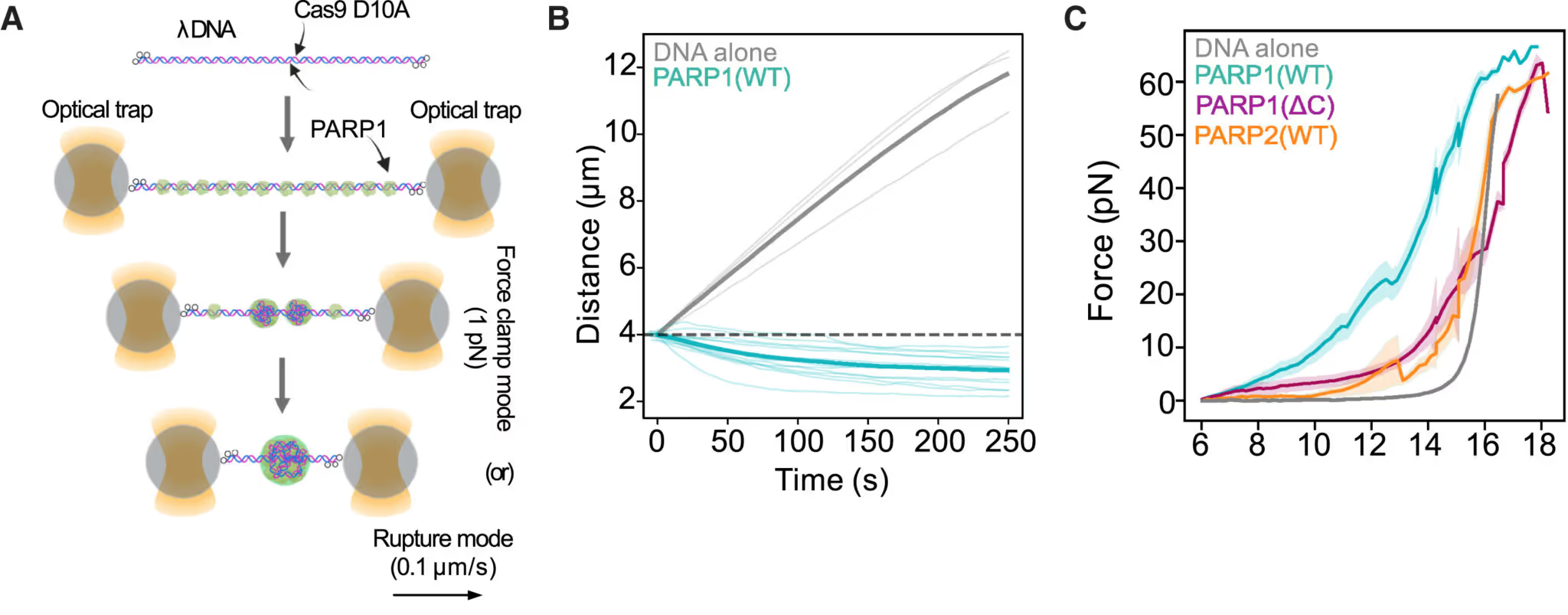
In the C-Trap, a single DNA molecule with nicks was held between two optical traps. Adding PARP1 to the assay causes PARP1 to form condensates at the DNA lesions (Figure A). A constant tension was applied on the DNA. When force is applied using the C-Trap, bare DNA extends like an elastic string (Figure B, grey line) while PARP1 mixed DNA shortens over time under the same force (Figure B, green line). This is direct evidence that PARP1-DNA condensates generate compaction forces which bring lesioned DNA together.
The C-Trap also allows scientists to stretch DNA, and precisely measure the force exerted. The force required to elongate DNA with PARP1 (WT) was much higher than for DNA alone or DNA with PARP1 (∆C) or PARP2 (Figure C) which don’t form condensates. Another direct evidence of PARP1 condensates physically connecting DNA lesions!
Scientific insights of this study
This study highlights PARP1 condensation’s critical role in DNA repair, utilizing C-Trap technology to unveil the dynamics of DNA repair processes in real-time. For the first time, researchers observed PARP1-DNA condensates actively securing and stabilizing broken strands of DNA, preventing them from separating. This breakthrough enhances our understanding of the molecular mechanisms protecting our genetic integrity.
Building on this, this research opens new pathways in healthcare, particularly in cancer treatment. With PARP1 inhibitors already in clinical use, deeper insights into PARP1 condensates could lead to innovative therapeutic strategies targeting DNA repair mechanisms, offering hope for advancements in treating cancer and other genetic diseases.
For further information, read the full article “PARP1-DNA co-condensation drives DNA repair site assembly to prevent disjunction of broken DNA ends”, published in Cell.
Are you interested in using dynamic single-molecule tools like the C-Trap for your research?
For more information, a demo, or a quote contact us!
Top and middle images adapted from Chappidi et al, Cell, 2024.