A recent publication in the journal Scientific Reports describes a new method to create arrays of nucleosome positioning motifs. Through sequential Gibson Assembly reactions, the researchers managed to clone a series of sequence motifs, with high affinity for histone octamers, into one DNA molecule.
The approach is ideal to start assessing nucleosome unwrapping and histone properties with dynamic single-molecule techniques, such as optical tweezers. In the study, they nicely show the method in action, using the C-Trap® with correlated optical tweezers and confocal fluorescence microscopy.
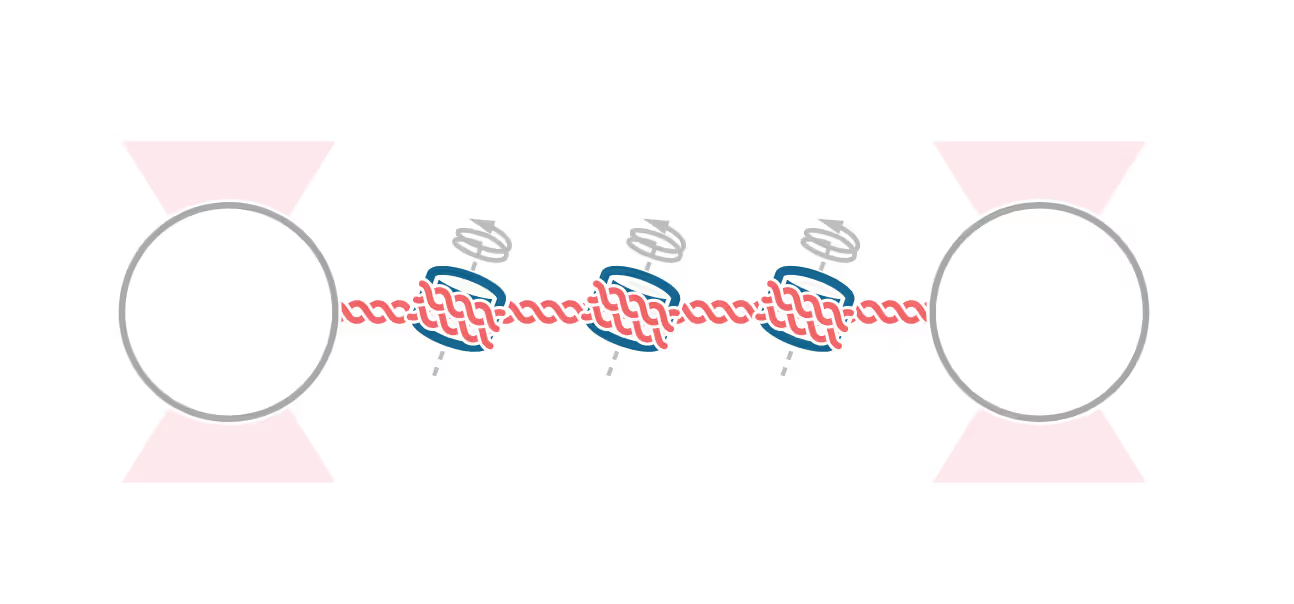
Through the Gibson Assembly molecular cloning method, Spakman et al. first managed to establish arrays of “601” motifs into a pKYB1 plasmid through parallel reactions. The 601 motif is an artificial DNA sequence with high affinity for histones and is commonly used to study nucleosome features.
The researchers tethered the linearized plasmid containing 12 motifs between two optically trapped beads and exposed it to fluorescently tagged nucleosomes through the C-Trap’s microfluidics system. By pulling the linearized plasmid at different forces, they could next assess and visualize the unwrapping properties of the reconstituted nucleosomes.
The combined optical tweezers and fluorescence imaging allowed the investigators to confirm the localization of histones under mechanical stress, which was previously unknown. Through this procedure, they found that histones remain bound to the array of nucleosome binding sites even at tensions that unwrap the DNA.
The study demonstrates an innovative strategy to create libraries of histone-binding sites, which can be used for different types of motifs. It contributes well to the single-molecule assessment of nucleosome properties as it provides a simple method to investigate and compare multiple binding sites in vitro.
Congratulations to Dian Spakman and Prof. Gijs Wuite at the Vrije Universiteit Amsterdam, and all the authors involved in this study!
For more information, read the full article published in the journal Scientific Reports titled “Constructing arrays of nucleosome positioning sequences using Gibson Assembly for single-molecule studies”.
Are you interested in using dynamic single-molecule tools like the C-Trap® for your research? Feel free to contact us for more information, a demo, or a quote.